-
Identify the Safest Python Version
Mar 6, 2025
There have been a numerous times when I would download and install Python using conda and known packages would fail to install because the Python version is too “new”. Thus, I’ve found the need to quickly identify a reasonably recent, stable, and “safe” version of Python to use and this is the command that I came up with it:
#!/bin/bash get_safe_python_version() { SAFE_PYTHON=`curl --location https://devguide.python.org/versions | xmllint --html --xpath '//section[@id="supported-versions"]//table/tbody/tr[count(//section[@id="supported-versions"]//table/tbody/tr[td[.="security"]]/preceding-sibling::*)]/td[1]/p/text()' - 2>/dev/null` } get_safe_python_version echo $SAFE_PYTHONEssentially, it scrapes the version table from “https://devguide.python.org/versions” and retrieves the penultimate security version (i.e., one version before the latest securty version).
As a bonus, you can also retrieve the minimum Python version or comptaible numpy/numba Python version using:
#!/bin/bash get_min__python_version() { MIN_PYTHON=`curl --location https://devguide.python.org/versions | xmllint --html --xpath '//section[@id="supported-versions"]//table/tbody/tr[last()]/td[1]/p/text()' - 2>/dev/null` } get_min_numba_numpy_version() { NUM_PYTHON=`curl --location https://numba.readthedocs.io/en/stable/user/installing.html#version-support-information | xmllint --html --xpath '//div[@id="version-support-information"]//table/tbody/tr[1]/td[3]/p/text()' - 2>/dev/null | awk '{print $1}' | sed s/\.x//`` }It’s certainly nothing fancy (and, admittedly, rather ugly) but it works like a charm!
-
Contributing to a Github PR
Jul 6, 2022
As an open source maintainer, I spend a good amount of time mentoring and providing feedback on pull requests (PRs). In some cases, providing comments to a PR may not be enough and I’ve had the occasional need to step in and make code contributions back to a contributor’s PR. How to accomplish this was a mystery to me but these steps seemed to work well:
- install the Github CLI (e.g.,
conda install -c conda-forge gh) - clone main (parent) repo (owned by me) (e.g., git clone https://github.com/TDAmeritrade/stumpy stumpy.git)
- navigate to the PR page and copy the Github CLI command provided in the
< > Codebutton in the upper right (e.g.,gh pr checkout 595) - navigate to the cloned repo in your terminal & execute the CLI cmd in Step 3
- finally, change the code (it will already be on the right branch), commit, and push
And that’s it! Thank you to all of those who chimed in with useful suggestions.
- install the Github CLI (e.g.,
-
Handling Special Characters In Your Proxy Password
Apr 24, 2022
In the past, I’ve discussed how not to fetch conda packages behind a firewall and, instead, how to leverage your coporate proxy. However, in any modern tech environment, you will likely be required to include special characters in your password that might cause your script to fail since special characters will need to be “URL encoded”. This post builds upon our previous knowledge and provides a solution to this problem:
#!/bin/bash urlencode(){ # Encodes spaces and non-alphanumeric characters str=$@ local length="${#str}" for (( i = 0; i < length; i++ )); do local c="${str:i:1}" case $c in [a-zA-Z0-9]) printf "$c" ;; *) printf '%%%02X' "'$c" esac done } proxy(){ hostname="proxy.yourcompany.com" port="8080" user="$(whoami)" read -s password password="$(urlencode $password)" export http_proxy=http://$user:$password@$hostname:$port export https_proxy=http://$user:$password@$hostname:$port export HTTP_PROXY=http://$user:$password@$hostname:$port export HTTPS_PROXY=https://$user:$password@$hostname:$port } proxy curl -O https://repo.anaconda.com/miniconda/Miniconda3-latest-MacOSX-x86_64.sh bash ./Miniconda3-latest-MacOSX-x86_64.sh -f -b -p miniconda3With the proxy function above, you can now update conda and install other packages with:
proxy conda update -y conda conda update -y --all conda install -y -c conda-forge stumpy numpy scipy numbaNow, you should be able to use the proxy and to install the software that you need to be productive!
-
Installing And Updating Conda With A Proxy
Nov 7, 2021
In the past, I’ve discussed how not to fetch conda packages behind a firewall. In this post, let’s assume that you are working behind a firewall in some corporate environment but your network administrator has given you access to a username/password-procted proxy:port that allows you to reach the “outside world”:
http://<username>:<password>@proxy.yourcompany.com:8080 https://<username>:<password>@proxy.yourcompany.com:8080
Now, how do we download, install, and updatecondabut make sure that we direct our requests through our proxy? First, let’s downloadminicondathrough our proxy and then install it:#!/bin/bash prox(){ export http_proxy=http://<username>:<password>@proxy.yourcompany.com:8080 export https_proxy=http://<username>:<password>@proxy.yourcompany.com:8080 export HTTP_PROXY=http://<username>:<password>@proxy.yourcompany.com:8080 export HTTPS_PROXY=https://<username>:<password>@proxy.yourcompany.com:8080 } proxy curl -O https://repo.anaconda.com/miniconda/Miniconda3-latest-MacOSX-x86_64.sh bash ./Miniconda3-latest-MacOSX-x86_64.sh -f -b -p miniconda3With the proxy function above, you can now update conda and install other packages with:
proxy conda update -y conda conda update -y --all conda install -y -c conda-forge stumpy numpy scipy numbaNow, you should be able to use the proxy and to install the software that you need to be productive!
-
Installing line_profiler For the Apple M1 ARM-based Chips
Apr 19, 2021
Recently, I found the need to install
line_profileron my Apple M1 machine in order to profile some code in Jupyter. Whilepipappeared to successfully install the package without any errors, I encountered segmentation faults when I loaded the package. Instead, the solution was to install directly from source :conda install -y -c conda-forge scikit-build cython git clone https://github.com/pyutils/line_profiler.git cd line_profiler && python setup.py install
Now, you should be able to useline_profiler!
-
Adding LaTeX Equations to Github Issues
Mar 31, 2021
Recently, I found the need to embed some LaTex equations in a Github issue and discovered that you can do this by using this HTML image tag along with the desired equation:
<img src="https://render.githubusercontent.com/render/math?math=a^{2} %2B b^{2} = c^{2}">
which should produce:
Now, you should be able to embed this anywhere!Check out this thread to add additional formatting like superscript or fractions
-
Installing Miniconda
Feb 19, 2021
When developing on a new VM, this is the basic setup that I like to build on top of:
#!/bin/bash MINICONDADIR="$HOME/miniconda3" rm -rf $MINICONDADIR wget -c https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh # curl -O 'https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh' bash ./Miniconda3-latest-Linux-x86_64.sh -f -b -p $MINICONDADIR $MINICONDADIR/bin/conda init $MINICONDADIR/bin/conda update -y conda $MINICONDADIR/bin/conda update -y --all # $MINICONDADIR/bin/conda install -y -c conda-forge python=3.9 $MINICONDADIR/bin/conda install -y -c conda-forge mamba jupyterlab if [[ `grep "alias python" $HOME/.bashrc | wc -l` -lt "1" ]]; then echo 'if [[ `which ipython | wc -l` -gt "0" ]]; then' >> $HOME/.bashrc echo ' alias python="ipython"' >> $HOME/.bashrc echo 'fi' >> $HOME/.bashrc fi if [[ `grep "alias ipynb" $HOME/.bashrc | wc -l` -lt "1" ]]; then echo 'if [[ `which jupyter-lab | wc -l` -gt "0" ]]; then' >> $HOME/.bashrc echo ' alias ipynb="jupyter-lab"' >> $HOME/.bashrc echo 'fi' >> $HOME/.bashrc fi if [[ `grep "alias rm" $HOME/.bashrc | wc -l` -lt "1" ]]; then echo 'alias rm="rm -i"' >> $HOME/.bashrc fi if [[ `grep "alias cp" $HOME/.bashrc | wc -l` -lt "1" ]]; then echo 'alias cp="cp -i"' >> $HOME/.bashrc fi if [[ `grep "alias vim" $HOME/.bashrc | wc -l` -lt "1" ]]; then echo 'alias vi="vim"' >> $HOME/.bashrc fi if [[ `grep "alias EDITOR" $HOME/.bashrc | wc -l` -lt "1" ]]; then echo 'export EDITOR="vim"' >> $HOME/.bashrc fi if [[ `grep syntax $HOME/.vimrc | wc -l` -lt "1" ]]; then echo "set viminfo=\'10,\"100,:20,%,n~/.viminfo" >> $HOME/.vimrc echo "" >> $HOME/.vimrc echo 'function! ResCur()' >> $HOME/.vimrc echo " if line(\"'\\\"\") <= line(\"$\")" >> $HOME/.vimrc echo " normal! g\`\"" >> $HOME/.vimrc echo ' return 1' >> $HOME/.vimrc echo ' endif' >> $HOME/.vimrc echo 'endfunction' >> $HOME/.vimrc echo "" >> $HOME/.vimrc echo 'augroup resCur' >> $HOME/.vimrc echo ' autocmd!' >> $HOME/.vimrc echo ' autocmd BufWinEnter * call ResCur()' >> $HOME/.vimrc echo 'augroup END' >> $HOME/.vimrc echo "" >> $HOME/.vimrc echo "set clipboard=unnamed" >> $HOME/.vimrc echo "set cindent" >> $HOME/.vimrc echo "set smartindent" >> $HOME/.vimrc echo "set autoindent" >> $HOME/.vimrc echo "set paste" >> $HOME/.vimrc echo "set ruler" >> $HOME/.vimrc echo "syntax on" >> $HOME/.vimrc echo "set tabstop=4" >> $HOME/.vimrc echo "set shiftwidth=4" >> $HOME/.vimrc echo "set expandtab" >> $HOME/.vimrc fi
And then, after installingminiconda, I just need to exit and re-enter the VM
-
Installing A Conda Environment
Jul 24, 2020
When developing STUMPY, I frequently and purposely delete my entire Python (conda) environment and start from scratch. This serves two purposes:
- It ensures that I continue developing and testing using the latest dependencies
- It helps to ensure that no new local dependencies have crept into STUMPY
However, every time I wipe out my environment (i.e.,
rm -rf /path/to/miniconda3/), I have to try and remember what I need to re-install. Instead, it’s far more efficient to document all of this in anenvironment.ymlfile:channels: - conda-forge - defaults dependencies: - python>=3.6 - numpy - scipy - numba - pandas - dask - distributed - coverage - flake8 - flake8-docstrings - black - pytest-cov - jupyterlab
And then, after reinstallingminiconda, I just need to execute the following commands in the same directory as theenvironment.ymlfileconda update -y conda conda update -y --all conda env update --file environment.yml
Note that this installs the desired packages in the base conda environment. To install this in a named environment, you’ll need:conda update -y conda conda update -y --all conda env update --name environment_name --file environment.yml
-
Finding the Top or Bottom K Elements from a NumPy Array
Jan 3, 2020
If you have a NumPy array, you can use
np.partitionto find the top k elements or the bottom k without having to sort the entire array first. To find the top k:top_k = np.partition(x, -k)[-k:] # Results are not sorted np.sort(tok_k)[::-1] # Sorted from largest to smallest
To find the bottom k:bottom_k = np.partition(x, k)[:k] # Results are not sorted np.sort(bottom_k) # Sorted from smallest to largest
Alternatively, you can usenp.argpartitionto find the indices of either the top k elements or the bottom k without having to sort the entire array first. To find the top k:idx = np.argpartition(x, -k)[-k:] # Indices not sorted idx[np.argsort(x[idx])][::-1] # Indices sorted by value from largest to smallest
To find the bottom k:idx = np.argpartition(x, k)[:k] # Indices not sorted idx[np.argsort(x[idx])] # Indices sorted by value from smallest to largest
-
Write and Read a NumPy Array
Dec 23, 2019
The documentation for writing/reading a NumPy array to/from a file without pickling is a tad hard to follow. One may need to do this when dealing with large NumPy arrays. Below is some simple code for writing and reading a NumPy array to a temporary file:
#!/usr/bin/env python import numpy as np import tempfile import os ftmp = tempfile.NamedTemporaryFile(delete=False) fname = ftmp.name + ".npy" inp = np.random.rand(100) np.save(fname, inp, allow_pickle=False) out = np.load(fname, allow_pickle=False) print(out) os.remove(fname)
Here, we leverage thetempfilemodule for dynamically creating our intermediate files and then deleting them only after the file is no longer needed. This is rather useful if you are using themultiprocessingmodule and your files are created within a subprocess. The key part to all of this is the necessity to append the.npyextension to the file name that is created bytempfileand also to turn off pickling (this will allow us to create files that are larger than 4GiB) in size.
-
Ignoring Requests SSL Verification in Third Party Apps
Dec 3, 2019
WARNING: This is unsafe!
One of the most annoying things is not being able to perform an https request using a third party application (e.g., twine) if you’re behind a network firewall that requires SSL verification. Luckily, you can turn this off by temporarily modifying the requests package. Open the
~/miniconda3/lib/python3.7/site-packages/requests/sessions.pyand, in theSessionclass, set#: SSL Verification default. self.verify = FalseAlternatively, directly from the command line, you might be able to tell the
sslPython module to temporarily disable SSL verification before running the Python program:PYTHONHTTPSVERIFY=0 python your_script.pyNote that this is a complete hack and not usually advised especially if the site that you are visiting is not trusted. This is only here for demonstration purposes. Do not use this code!
-
Fetching Code Repositories Without SSL
Nov 6, 2019
One of the most annoying things is not being able to clone a code repository from trusted sources such as Github or Bitbucket if you’re behind a network firewall that requires SSL verification. Luckily, you can turn this off in Git via:
git config --global http.sslverify falseIf code (or a file) is retrieved via
wgetthen you can turn off certificate checking globally with:check_certificate = off
-
Creating a Time-Dependent Sleep Function
Oct 23, 2019
I’ve been retrieving data from a database that is being accessed by many people and it is especially busy during the mornings. So, to avoid this congestion, I set a simple timer in my Python code that detects the (Eastern) time and sleeps for a minute until it is outside of the congested hours. Here’s what it looks like:
import datetime import time import pytz # Sleep between 9a-11a Eastern Time while 9 <= datetime.datetime.now(pytz.timezone('America/New_York')).time().hour < 11: time.sleep(60)
Things seemed fine when I ran the script locally and it would update the cron job as I had expected. So, I let it go on its merry way. A couple of weeks later and the site that I was monitoring goes down but I don’t get any notifications. Again, running the above code directly from the command line worked perfectly but the cron job was not doing anything. Upon further inspection, the cron job was somehow looking for therun_monitor.shscript in thehomedirectory. Then it dawned on me that thepwdwas the problem. Since the cron job is being executed relative to the home directory, thepwdcommand within the script actually returns the home directory and not the location of therun_monitor.shscript. So, instead, we need the$APPDIRto point to the location of therun_monitor.shscript like this:#!/bin/sh APPDIR="$( cd "$( dirname "$0" )" && pwd )" HTTP_RESPONSE=`curl -k -sL -w "%{http_code}" "http://www.google.com" -o /dev/null` if [ ! $HTTP_RESPONSE -eq 200 ]; then # Replace this line to send a message to channel # Remove/add/update cron job to once every hour (crontab -l | grep -v "$APPDIR/run_monitor.sh"; echo "0 * * * * $APPDIR/run_monitor.sh") | sort - | uniq - | crontab - else # Remove/add/update cron job to once every minute (crontab -l | grep -v "$APPDIR/run_monitor.sh"; echo "* * * * * $APPDIR/run_monitor.sh") | sort - | uniq - | crontab -
Problem solved! It’s a subtle bug that was hard to detect/debug but became obvious only after I spent some time thinking about it. I hope to never encounter this again!
-
Special Cron Job
Oct 16, 2019
Today, I encountered a really nasty scripting “bug” that involves running a cron job. I have a simple script called
run_monitor.shthat:- Curls a website to get its response status
- Checks the response status
- Updates the cron job depending on the status
#!/bin/sh APPDIR=`pwd` HTTP_RESPONSE=`curl -k -sL -w "%{http_code}" "http://www.google.com" -o /dev/null` if [ ! $HTTP_RESPONSE -eq 200 ]; then # Replace this line to send a message to channel # Remove/add/update cron job to once every hour (crontab -l | grep -v "$APPDIR/run_monitor.sh"; echo "0 * * * * $APPDIR/run_monitor.sh") | sort - | uniq - | crontab - else # Remove/add/update cron job to once every minute (crontab -l | grep -v "$APPDIR/run_monitor.sh"; echo "* * * * * $APPDIR/run_monitor.sh") | sort - | uniq - | crontab -
Things seemed fine when I ran the script locally and it would update the cron job as I had expected. So, I let it go on its merry way. A couple of weeks later and the site that I was monitoring goes down but I don’t get any notifications. Again, running the above code directly from the command line worked perfectly but the cron job was not doing anything. Upon further inspection, the cron job was somehow looking for therun_monitor.shscript in thehomedirectory. Then it dawned on me that thepwdwas the problem. Since the cron job is being executed relative to the home directory, thepwdcommand within the script actually returns the home directory and not the location of therun_monitor.shscript. So, instead, we need the$APPDIRto point to the location of therun_monitor.shscript like this:#!/bin/sh APPDIR="$( cd "$( dirname "$0" )" && pwd )" HTTP_RESPONSE=`curl -k -sL -w "%{http_code}" "http://www.google.com" -o /dev/null` if [ ! $HTTP_RESPONSE -eq 200 ]; then # Replace this line to send a message to channel # Remove/add/update cron job to once every hour (crontab -l | grep -v "$APPDIR/run_monitor.sh"; echo "0 * * * * $APPDIR/run_monitor.sh") | sort - | uniq - | crontab - else # Remove/add/update cron job to once every minute (crontab -l | grep -v "$APPDIR/run_monitor.sh"; echo "* * * * * $APPDIR/run_monitor.sh") | sort - | uniq - | crontab -
Problem solved! It’s a subtle bug that was hard to detect/debug but became obvious only after I spent some time thinking about it. I hope to never encounter this again!
-
Detecting File Encodings
Oct 16, 2019
There have been numerous times where I’ve tried read a CSV file into a Pandas DataFrame and it fails due to the file encoding. The best thing to do is to detect the file encoding by reading a few lines from the file and then passing that encoding to Pandas. The file encoding detection part can be done with the
chardetpackage and below is a convenience function for grabbing the encoding for the firstn_lines:from chardet.universaldetector import UniversalDetector def get_encoding(fname, n_lines=50): detector = UniversalDetector() detector.reset() with open(fname, 'rb') as f: for i, row in enumerate(f): detector.feed(row) if i >= n_lines-1 or detector.done: break detector.close() return detector.result['encoding']
Then you can call this function with a file name:encoding = get_encoding("file.csv") df = pd.read_csv("file.csv", encoding=encoding)
I hope this help you too!
-
Checking if a Local Dask Cluster is Running
Oct 8, 2019
One thing that I find myself doing fairly often is spinning up a local Dask cluster, forgetting that it is running, and then starting it up again on a different port. This is here as a reminder that you can first
tryto connect to the cluster and, if it times out, then create theLocalClusteron the fly:from distributed import Client, LocalCluster try: client = Client('tcp://localhost:8786', timeout='2s') except OSError: cluster = LocalCluster(scheduler_port=8786) client = Client(cluster) client.restart() client
Since I always forget how to do this, I hope that others will find this helpful!
-
Conda Base Ignores Linux .Profile
Aug 5, 2019
Recently, I reinstalled miniconda and allowed it to set up
conda initon Mac OSX. From what I could tell, it:- Updates
.bash_profileand checks to see ifconda.shexists and executes it - Otherwise, if 1. is False, then pre-pend the
miniconda3/bindirectory to$PATH - Add
. /Users/law978/miniconda3/etc/profile.d/conda.shto the.profileand remove references to pre-pendminiconda3/binto$PATH
Now, this seemed great but I noticed that all of the aliases that had been set in
.profilewere being ignored. In fact, it looks like.profilewasn’t being sourced at all. When I looked inside of.bash_profile, I realized that the usual command to source the contents of.profilewas missing:if [ -f ~/.profile ]; then source ~/.profile fi
Simply adding these three lines back to.bash_profiledid the trick. So, now, all of my aliases are loaded when a new terminal is opened andcondais initialized according to plan.
- Updates
-
Leveraging Sparse Arrays for Large-ish (Feature) Data Sets
Jul 26, 2019
When building a machine learning model, you’ll often have your feature data stored in one or more database tables (e.g., with a couple of million rows and maybe a thousand columns) where each row represents, say, an individual user and each column represents some feature that you’ve engineered (i.e., number of logins to your site, number of daily purchases, number of ads clicked on, 52 consecutive columns storing a weekly moving average over a year, etc). Since storage is cheap and you can’t afford to pay for a compute cluster, your instinct might be to download the full table onto your laptop as a quick and dirty CSV file or, possibly, in a more portable and compact parquet format. However, depending on your local computer hardware, trying to load that full data set into a Pandas DataFrame might consume all of your memory and cause your system to grind to a halt!
Luckily, there is one approach that a lot of people often overlook but that might work wonderfully for you. The secret is to exploit the inherent sparsity that likely exists within your large data set!
-
A Simple Restart Script
Jul 20, 2019
Let’s say that you have a Python script (or other command), called run.py, that you’d like to run in the background on a Linux OS and then end the session:
import time while True: time.sleep(5) print("I'm awake!")
To prevent the session from timing out, one common way to avoid this is to use:nohup ./run.py &> LOG &
This will execute the command in the background and also write any output to a LOG file. However, if I come back months later and forget that it is still running and I execute this command again then I might end up having multiple processes running the same command. To avoid this, we can do a little better and ensure that we kill the existing process. This typically requires recording the PID of the original process:cat PID | xargs kill -9 2>/dev/null nohup ./run.py &> LOG & echo $! > PID
This simple little trick has come in handy many times and I hope you find it useful too!
-
STUMPY - A Re-newed Approach to Time Series Analysis
May 13, 2019

Thanks to the support of TD Ameritrade, I recently open sourced (BSD-3-Clause) a new, powerful, and scalable Python library called STUMPY that can be used for a variety of time series data mining tasks. At the heart of it, this library takes any time series or sequential data and efficiently computes something called the matrix profile, which, with only a few extra lines of code, enables you to perform:
- pattern/motif (approximately repeated subsequences within a longer time series) discovery
- anomaly/novelty (discord) discovery
- shapelet discovery
- semantic segmentation
- density estimation
- time series chains (temporally ordered set of subsequence patterns)
- and more…
First, let’s install stumpy with Conda (preferred):conda install -c conda-forge stumpy
or, alternatively, you can install stumpy with Pip:pip install stumpy
Once stumpy is installed, typical usage would be to take your time series and compute the matrix profile:import stumpy import numpy as np your_time_series = np.random.rand(10000) window_size = 50 # Approximately, how many data points might be found in a pattern matrix_profile = stumpy.stump(your_time_series, m=window_size)
For a more detailed example, check out our tutorials and documentation or feel free to file a Github issue. We welcome contributions in any form!
I’d love to hear from you so let me know what you think!
-
Discussing Data Science R&D on the DataFramed Podcast
Apr 1, 2019

Due to DataCamp’s internal mishandling of the sexual assault, I will no longer be promoting this podcast recording and, instead, encourage you to read more about what happened here.
I was recently invited to sit down and chat with Hugo Bowne-Anderson on the DataFramed podcast to talk Data Science R&D. Have a listen and leave your comments below!
-
Setting Values of a Sparse Matrix
Feb 27, 2019
Let’s say that you have a sparse matrix:
import numpy as np from scipy.sparse import x = csr_matrix(np.array([[1, 0, 2, 0, 3], [0, 4, 0, 5, 0]])) print(x)<2x5 sparse matrix of type '<class 'numpy.int64'>' with 5 stored elements in Compressed Sparse Row format>
One of the most common things that you might want to do is to make a conditional selection from the matrix and then set those particular elements of the matrix to, say, zero. For example, we can take our matrix from above and set all elements that have a value that are less than three to zero. Naively, one could do:x[x < 3] = 0
This works and is fine for small matrices. However, you’ll likely encounter a warning message such as the following:/home/miniconda3/lib/python3.6/site-packages/scipy/sparse/compressed.py:282: SparseEfficiencyWarning: Comparing a sparse matrix with a scalar greater than zero using < is inefficient, try using >= instead. warn(bad_scalar_msg, SparseEfficiencyWarning)
The problem here is that for large sparse matrices, the majority of the matrix is full of zeros and so the<comparison becomes highly inefficient. Instead, you really only want to perform your comparison only with the nonzero elements of the matrix. However, this takes a little more work and a few more lines of code to accomplish the same thing. Additionally, we want to avoid converting our sparse matrices into costly dense arrays.
-
Pip Installing Wheels with Conda GCC/G++
Jan 17, 2019
I was trying to
pip installa simple package that contained wheels that needed to be compiled with both GCC and G++. Of course, without using SUDO (i.e.,yum install gcc) meant that I needed to rely on my good friend, Conda:conda install gcc_linux-64 conda install gxx_linux-64
Now, we aren’t done yet! According to the conda documentation, the compilers are found in/path/to/anaconda/binbut thegccandg++executables are prefixed with something likex86_64-conda-cos6-linux-gnu-gcc. So, we’ll need to create some symbolic links to these executables:ln -s /path/to/anaconda/bin/x86_64-conda-cos6-linux-gnu-gcc /path/to/anaconda/bin/gcc ln -s /path/to/anaconda/bin/x86_64-conda-cos6-linux-gnu-g++ /path/to/anaconda/bin/g++
Alternatively, you can avoid the symbolic link by setting your environment variables accordinglyexport CC=/path/to/anaconda/bin/x86_64-conda_cos6-linux-gnu-gcc export CXX=/path/to/anaconda/bin/x86_64-conda_cos6-linux-gnu-g++
Now, my pip install command is able to compile the wheel successfully!
-
Compiling Facebook's StarSpace with Conda Boost
Jun 17, 2018
Recently, I was playing around with Facebook’s StarSpace, a general-purpose neural model for efficient learning of entity embeddings for solving a wide variety of problems. According to the installation instructions, you need a C++11 compiler and the Boost library. I already had GCC installed and Boost was only a quick conda command away:
conda install boost
The StarSpace Makefile is hardcoded to look for the Boost library in /usr/local/bin/boost_1_63_0/, which is a problem. But how should I modify the StarSpace Makefile so that it knew where to include the Boost library? After a little digging, I found the Boost files in/path/to/anaconda/include. So, all I had to do was modify the following line in the StarSpace Makefile:#BOOST_DIR = /usr/local/bin/boost_1_63_0/ BOOST_DIR = /path/to/anaconda/include/
Executedmakeon the command line and everything compiled nicely! Yay!
-
I'm Melting! From Wide to Long Format and Quarterly Groupby
Dec 27, 2017
Recently, a colleague of mine asked me how one might go about taking a dataset that is in wide format and converting it into long format so that you could then perform some groupby operations by quarter.
Here’s a quick example to illustrate one way to go about this using the Pandas melt function.Getting Started
Let’s import the Pandas packageimport pandas as pdLoad Some Data
First, we’ll create a fake dataframe that contains the name of a state and city along with some data for each month in the year 2000. For simplicity, imagine that the data are the number of Canadians spotted eating poutine.df = pd.DataFrame([['NY', 'New York', 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13], ['MI', 'Ann Arbor', 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17], ['OR', 'Portland', 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21], ], columns=['state', 'city', '2000-01', '2000-02', '2000-03', '2000-04', '2000-05', '2000-06', '2000-07', '2000-08', '2000-09', '2000-10', '2000-11', '2000-12', '2001-01' ]) df
-
Select Rows with Keys Matching Multiple Columns in Subquery
Oct 17, 2016
When you query a database table using SQL, you might find the need to:
- select rows from table A using a certain criteria (i.e., a WHERE clause)
- then, use one or more columns from result set (coming from the above query)
as a subquery to subselect from table B
You can do this quite easily in SQL
import pandas as pd from pandasql import sqldf # pip install pandasql from Yhatdf_vals = pd.DataFrame({'key1': ['A', 'A','C', 'E', 'G'], 'key2': ['B', 'Z', 'D', 'F', 'H'], 'val': ['2','3','4','5','6']}) df_valskey1 key2 val 0 A B 2 1 A Z 3 2 C D 4 3 E F 5 4 G H 6 df_colors = pd.DataFrame({'key1': ['A', 'A','C', 'E', 'G'], 'key2': ['B', 'Z', 'D', 'F', 'H'], 'color': ['red','orange','yellow','green','blue']}) df_colorscolor key1 key2 0 red A B 1 orange A Z 2 yellow C D 3 green E F 4 blue G H
So, if we wanted to grab all rows from df_colors where the value in df_vals is inclusively between 2 and 6, then:
-
Pandas Split-Apply-Combine Example
May 28, 2016
There are times when I want to use split-apply-combine to save the results of a groupby to a json file while preserving the resulting column values as a list. Before we start, let’s import Pandas and generate a dataframe with some example email data
Import Pandas and Create an Email DataFrame
import pandas as pd import numpy as npdf = pd.DataFrame({'Sender': ['Alice', 'Alice', 'Bob', 'Carl', 'Bob', 'Alice'], 'Receiver': ['David', 'Eric', 'Frank', 'Ginger', 'Holly', 'Ingrid'], 'Emails': [9, 3, 5, 1, 6, 7] }) df
-
Pandas End-to-End Example
May 25, 2016
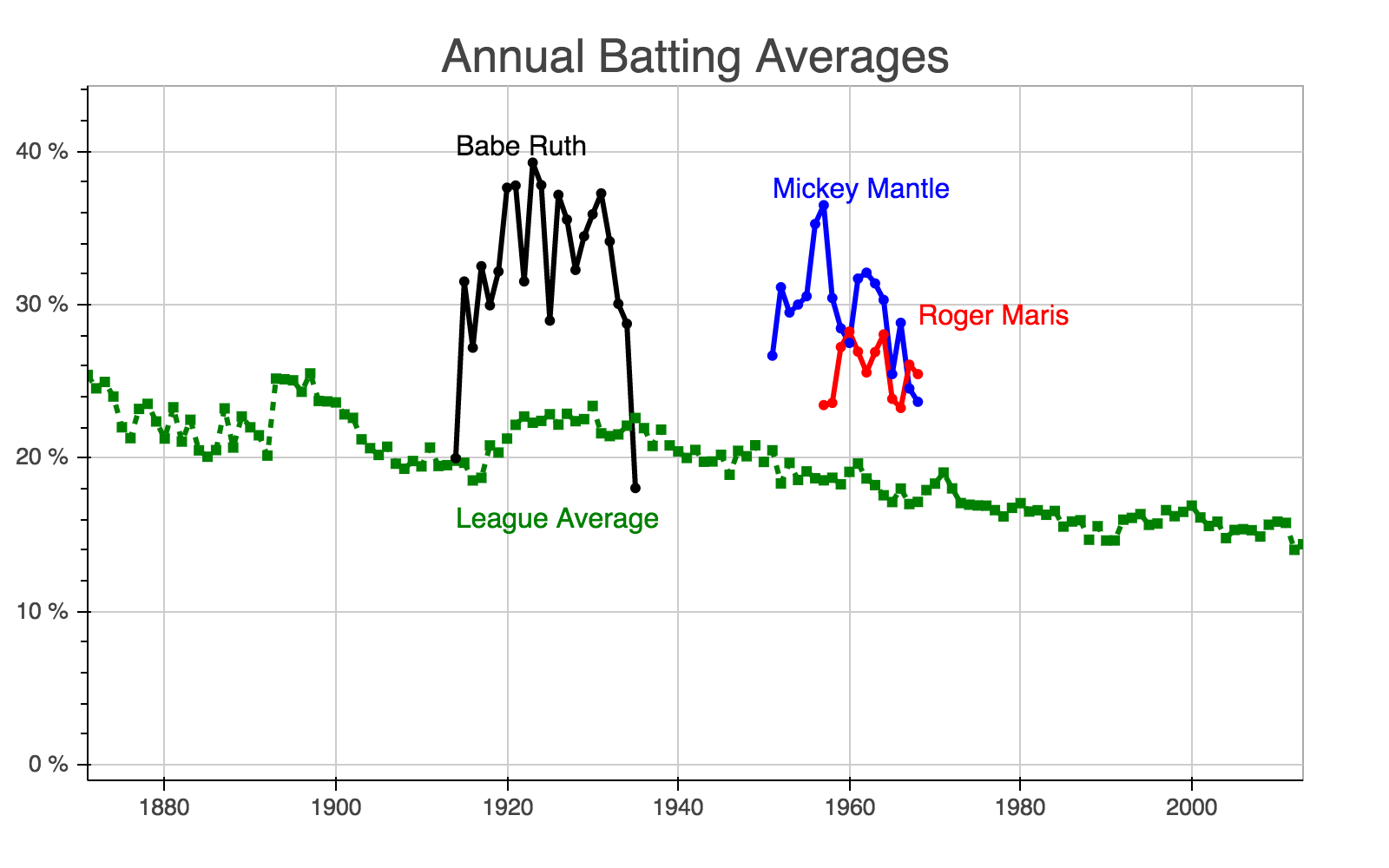
The indexing capabilities that come with Pandas are incredibly useful. However, I find myself forgetting the concepts beyond the basics when I haven’t touched Pandas in a while. This tutorial serves as my own personal reminder but I hope others will find it helpful as well.
To motivate this, we we’ll explore a baseball dataset and plot batting averages for some of the greatest players of all time.
-
Fetching Conda Packages Behind a Firewall
Dec 23, 2015
WARNING: This is unsafe!
One of the most annoying things is not being able to update software if you’re behind a network firewall that requires SSL verification. You can turn this off in Anaconda via
conda config --set ssl_verify no
and for pip viapip install --trusted-host pypi.python.org --trusted-host pypi.org --trusted-host files.pythonhosted.org <package name>
Optionally, you can also specify the package version like this:pip install --trusted-host pypi.python.org --trusted-host pypi.org --trusted-host files.pythonhosted.org <package name>[=0.1.2]
Better yet, you can permanently set the trusted-host by adding the following to the $HOME/.pip/pip.conf file:[global] trusted-host = pypi.python.org files.pythonhosted.org pypi.orgAlternatively, you can also temporarily disable SSL verification from the command line with:
PYTHONHTTPSVERIFY=0 pip install some_trusted_package_nameDo not use this code! –>
-
Convert a Pandas DataFrame to Numeric
Dec 15, 2015
Pandas has deprecated the use of
convert_objectto convert a dataframe into, say, float or datetime. Instead, for a series, one should use:df['A'] = df['A'].to_numeric()
or, for an entire dataframe:df = df.apply(to_numeric)
-
Python 2 Unicode Problem
Dec 1, 2015

The following Python error is one of the most annoying one’s I’ve ever encountered:
Traceback (most recent call last): File "./test.py", line 3, in <module> print out UnicodeEncodeError: 'ascii' codec can't encode character u'\u2019' in position 0: ordinal not in range(128)
Essentially, you can’t write unicode characters as string unless you’ve converted the text to a string first before printing it. A detailed explanation can be found in Kumar McMillan’s wonderful talk titled ‘Unicode in Python, Completely Demystified’. To summarize, McMillan offers three useful yet simple rules:
-
2015 Nobel Prize in Chemistry Awarded to DNA Repair
Oct 7, 2015
Today, the 2015 Nobel Prize in Chemistry was awarded to the field of DNA repair. I am especially excited by this news since I had spent six years researching the role that DNA base-flipping plays in DNA repair when I was a graduate student studying at Michigan State University under the mentorship of Dr. Michael Feig. Thus, my research sat at the crossroads between the exciting worlds of computational chemistry (which was awarded the Nobel Prize in Chemistry two years ago in 2013) and DNA repair which have ultimately shaped my appreciation for doing science.
Dr. Paul Modrich, one of the three Nobel Prize recipients this year, is a pioneer in the field of DNA mismatch repair and has spent decades trying to understand the mechanism by which humans (and other eukaryotes) maintain the efficacy and fidelity of their genome. As a computational biochemist/biophysicist, I am honored to have had the opportunity to make significant contributions to this field of research and am delighted to see this area be recognized!
Others scientists who have also made an impact in the area of DNA mismatch repair include (in no particular order) Drs. Richard Kolodner, Richard Fishel, Thomas Kunkel, Dorothy Erie, Manju Hingorani, Peggy Hsieh, Shayantani Mukherjee, Alexander Predeus, Meindert Lamers, Titia Sixma, et al.
Congratulations to all!
-
Installing Downloaded Anaconda Python Packages
Sep 22, 2015

If you work in a secure network at your job then conda may not be able to hit the Anaconda repositories directly even if it’s for accessing free packages. Additionally, it’s not recommended to use pip over conda when installing new packages. However, installing new packages can be done manually by:
- Downloading the package(s) (and its necessary dependencies) directly from the Continuum Repo
- And installing the tar.bz2 file using
conda install ./package_name.tar.bz2
-
Using NumPy Argmin or Argmax Along with a Conditional
Sep 10, 2015

It’s no secret that I love me some Python! Yes, even more than Perl, my first love from my graduate school days.
I’ve always found NumPy to be great for manipulating, analyzing, or transforming arrays containing large numerical data sets. It is both fast and efficient and it comes with a tonne of great functions.
-
Modern Data Scientist
Aug 8, 2015
The qualities of a modern data scientist is summed up very nicely in this article/guide and image by Marketing Distillery. As they point out, the team should be composed of people with a “mixture of broad, multidisciplinary skills ranging from an intersection of mathematics, statistics, computer science, communication and business”. More importantly:
“Being a data scientist is not only about data crunching. It’s about understanding the business challenge, creating some valuable actionable insights to the data, and communicating their findings to the business”.
I couldn’t agree more!
-
Anaconda Environment
Jul 9, 2015

I’ve been using Continuum’s enterprise Python distribution package, Anaconda, for several months now and I love it. Recently, people have been asking about Python 2.7 vs Python 3.x and so I looked into how to switch between these environments using Anaconda.
In fact, it’s quite straightforward and painless with Anaconda.
To set up a new environment with Python 3.4:
-
Stitch Fix Loves UNIX
May 27, 2015
The wonderful group of people at Stitch Fix has shared an informative list of useful UNIX commands. Go check it out now!
-
Drafts in Jekyll
Mar 14, 2015
The great thing about Jekyll is that you can start writing a draft without publishing it and still be able to see the post locally.
- Create a draft directory called
_draftsin the root directory - Create a new post in this directory but omit the date in the file name
- Serve up the page locally using
jekyll serve --drafts
Then, Jekyll will automatically adjust the date of the post to today’s date and display the post as the most recent post. Note that this post won’t be displayed on your github pages since they aren’t using the
--draftsoption. So, you’ll be able to save all of your drafts without worrying about them showing up on your live site. Once the post is ready for the prime time, then simply move it over to the_postsdirectory and prepend a date to the file name. That’s it!
- Create a draft directory called
-
3D Coordinates Represented on a 2D Triangle
Mar 6, 2015
I came across this interesting way of showing 3D coordinates on a 2D triangle published in the Journal of Physical Chemistry B. It takes a minute to orient yourself and figure out how best to interpret the results but the idea is pretty cool. I wonder what type of geometric transformation is need to create this plot. Once I figure it out, I’ll be sure to blog about it and prototype it in Python!
-
MathJax
Feb 28, 2015
As I embark on the PyESL project, I’ll need to include math equations in future blog posts. The easiest way to accomplish this is to use MathJax so that I can incorporate Tex/LaTeX/MathML-based equations within HTML. In Jekyll, all you need to do is add the MathJax javascript to the header section of your default.html and add a new variable to your _config.yml file.
<head> ... <script type="text/javascript" src="http://cdn.mathjax.org/mathjax/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML"> </script> ... </head>
and add the following to your _config.yml file:markdown: kramdown
For example, this markdown:Inline equation \\( {y} = {m}{x} + {b} \\) and block equation \\[ {y} = {m}{x}+{b} \\]
produces:Inline equation \( {y} = {m}{x}+{b} \) and block equation \[ {y} = {m}{x}+{b} \]
Here, the parentheses denote an inline equation while the square brackets denote a block equation.
And this is a multiline equation:
-
Tag Aware Previous/Next Links for Jekyll
Feb 22, 2015
Creating and maintaining a vanilla Jekyll-Boostrap website is pretty straightforward. However, I couldn’t find an obvious way to customize the previous/next links below each blog post so that:
- The links were aware of the tags listed in the front matter
- The method did not depend on plugin (since my site is being hosted on Github)
After tonnes of digging, I managed to piece together a Liquid-based solution (see my last post, to see how I add Liquid code in Jekyll)!
-
Escaping Liquid Code in Jekyll
Feb 21, 2015
To document some of my challenges in customizing this site, I’ve had to delve into Liquid code. However, adding Liquid code tags in Jekyll can be quite tricky and painful. Luckily, some smart people have identified a couple of nice solutions exists. Below is the markdown code that I’ve adopted for use in future posts:
{% highlight html %}{% raw %} \\Place Liquid and HTML code here {% endraw %}{% endhighlight %}
-
Elements of Statistical Learning
Feb 20, 2015

My new book purchase, Elements of Statistical Learning by Trevor Hastie, Robert Tibshirani, and Jerome Friedman arrived in the mail the last week and I’m excited to get reading! Springer was also kind enough to make this classic book available free to download. Get your copy here! Python implementations of each chapter will be added in the PyESL section.
-
Github and Jekyll-Bootstrap, FTW!
Feb 17, 2015
My blog is finally up and running! Currently, it’s being hosted (for free) and backed up on Github pages using vanilla Jekyll-Bootstrap. Font awesome, which definitely lives up to its name, was used to produce the social icons along the navigation bar and Dropbox is being used for redundancy. More design customizations will follow but I’m loving how easy the process has been!



